A Brief History of Advanced Normalization Tools (ANTs)
Brian B. Avants (PENN) and
Nicholas J. Tustison (UVA)

This talk is online at http://stnava.github.io/ANTsTalk/ with colored links meant to be clicked for more information
Background
Image mapping & perception: 1878

Francis Galton: Can we see criminality in the face?
(maybe he should have used ANTs?)
Founding developers

Long-term collaborators

\(+\) neurodebian, slicer, brainsfit, nipype, itk and more …
General purpose library for multivariate image registration, segmentation & statistical analysis tools
170,000+ lines of C++, 6\(+\) years of work, 15+ collaborators.
Generic mathematical methods that are tunable for application specific domains: no-free lunch
Deep testing on multiple platforms … osx, linux, windows.
Several “wins” in public knock-abouts ( Klein 2009, Murphy 2011, SATA 2012 and 2013, BRATS 2013, others )
An algorithm must use prior knowledge about a problem
to do well on that problem ANTs optimizes mathematically well-defined objective functions guided by prior knowledge …
… including that of developers, domain experts and other colleagues …
plug your ideas into our software to gain insight into biomedical data …
our strong mathematical and software engineering foundation leads to near limitless opportunities for innovation in a variety of application domains
ANTs is open to different image types, multiple modalities, anatomical regions, segmentation priors, etc.
ANTs & Neuroscience
We need statistical image analysis
at several scales in modern neuroscience
Macro: in vivo structural and functional MRI
Micro: high-resolution post-mortem MRI links with in vivo MRI
Nano: neuron reconstruction …
Solutions that are consistent across these scales have the potential to build multi-scale feature sets or templates and provide new insights into brain structure and function
E.g. Parcellation constraints based on histology, tractography, function …
Statistical definitions of anatomy/pathology?
Reinvention of these solutions within each lab … can we mitigate this?
Reduce, reuse, recycle …
ANTs Lineage
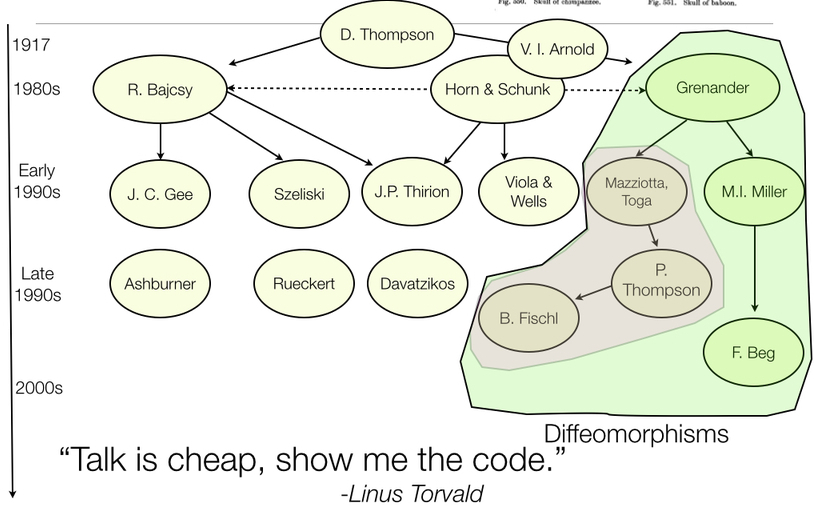
References: Horn and Schunck (1981), Gee, Reivich, and Bajcsy (1993), Grenander (1993), Thompson et al. (2001), Miller, Trouve, and Younes (2002), Shen and Davatzikos (2002), Arnold (2014), Thirion (1998), Rueckert et al. (1999), Fischl (2012), Ashburner (2012)
Diffeomorphisms
plausible physical modeling of large, invertible deformations
“differentiable map with differentiable inverse”
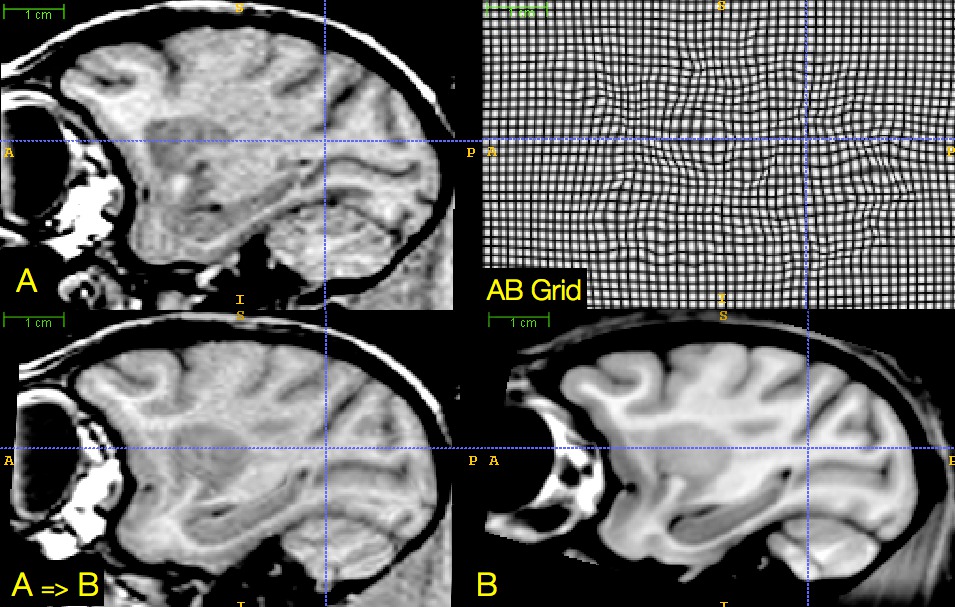
Fine-grained and flexible maps
ANTs: Beyond Registration
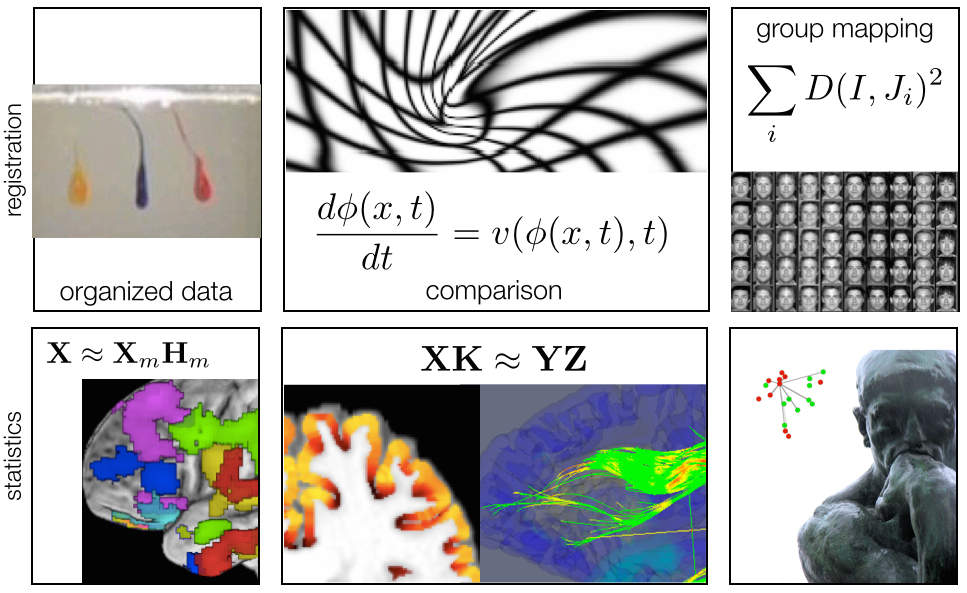
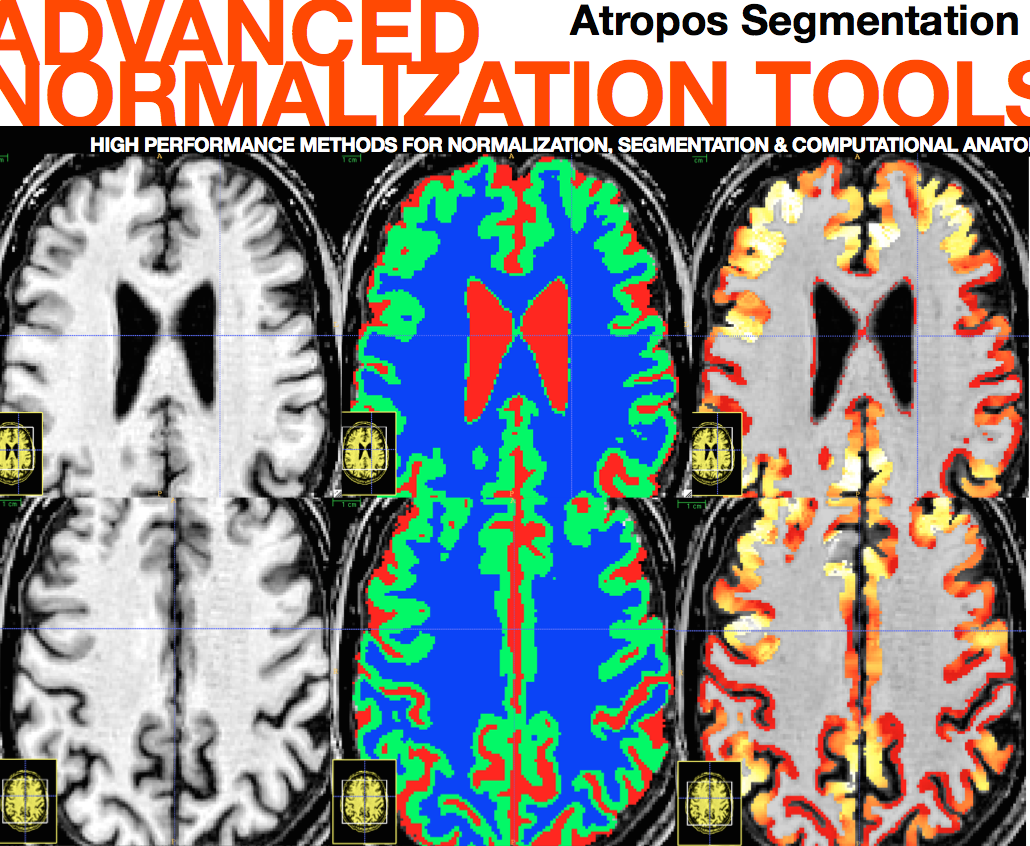
Atropos segmentation, N4 inhomogeneity correction, Eigenanatomy, SCCAN, Prior-constrained PCA, and atlas-based label fusion and MALF (powerful expert systems for segmentation)
Definitions
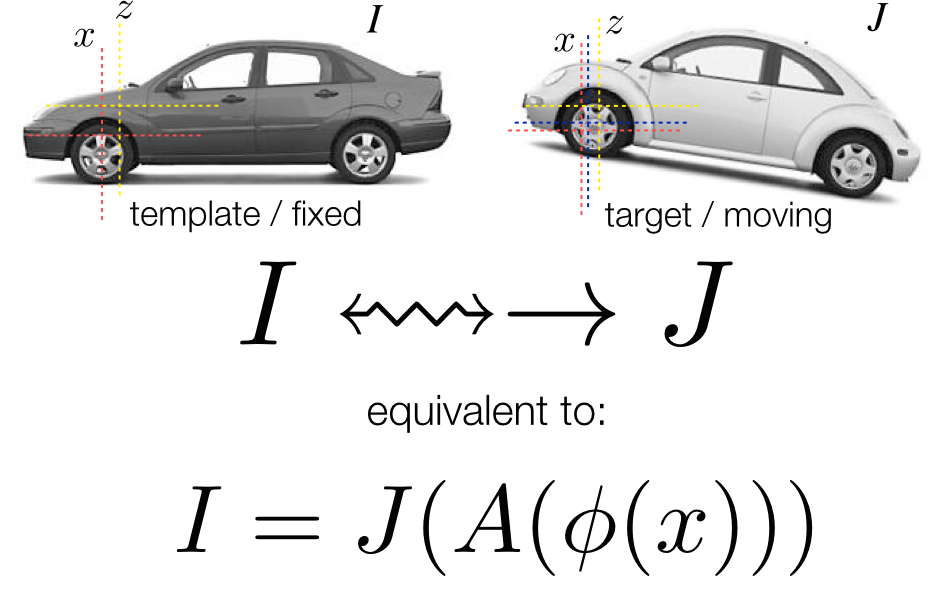
Registration \(=\) estimate an “optimal” geometric mapping between image pairs or image sets (e.g. Affine)
Similarity \(=\) a function relating one image to another, given a transformation (e.g. mutual information)
Diffeomorphisms \(=\) differentiable map with differentiable inverse (e.g. “silly putty”, viscous fluid)
Segmentation \(=\) labeling tissue or anatomy in images, usually automated (e.g. K-means)
Multivariate \(=\) using many voxels or measurements at once (e.g. PCA, \(p >> n\) ridge regression)
Multiple modality \(=\) using many modalities at once (e.g. DTI and T1 and BOLD)
MALF: multi-atlas label fusion - using anatomical dictionaries to label new data
Solutions to challenging statistical image processing problems usually need elements from each of the above
Medical Image Registration
Fundamental tool for
morphometry, segmentation,
motion estimation and
data cleaning
we can compare
apples and oranges …
apples and oranges …

initialization
apples and oranges …

RGB affine
apples and oranges …

RGB deformable registration - i.e. registration on color
The Technical Framework
… and most of it multivariate.
ANTs Nomenclature / Standards
ANTs Nomenclature / Standards
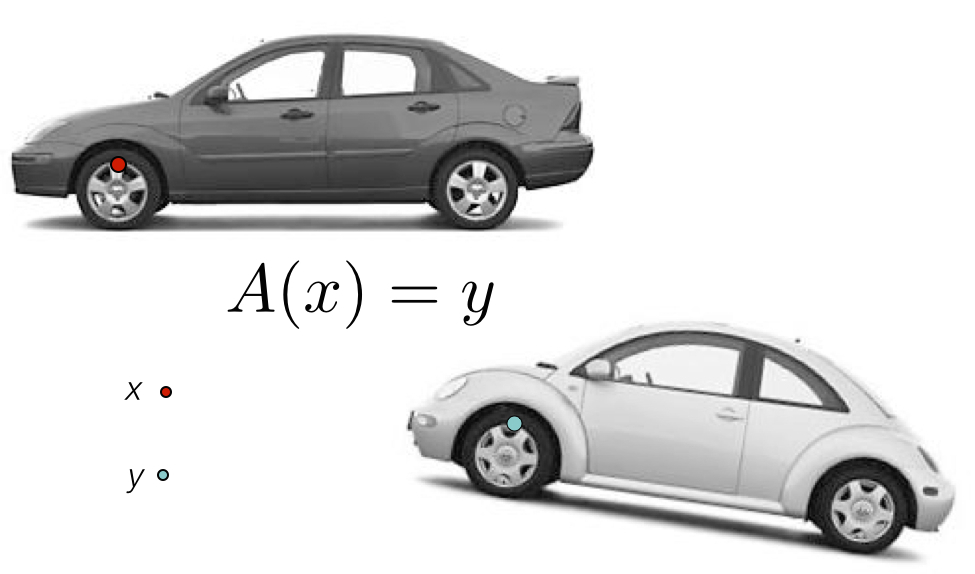
ANTs Nomenclature / Standards
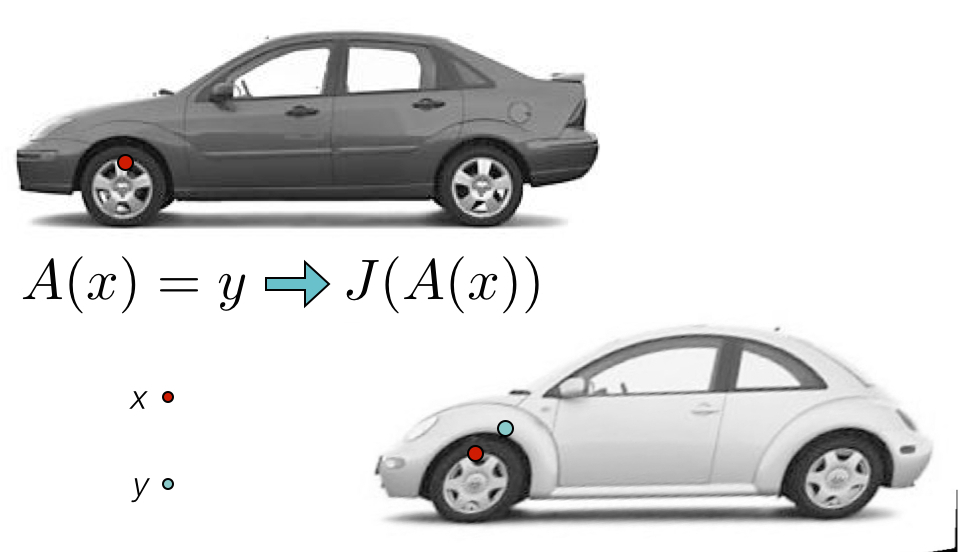
ANTs Nomenclature / Standards

The optimization problem
Find mapping \[ \color{red}{ \phi(x,p) \in \mathcal{T} }\] such that
\[ \color{red}{ M(I,J,\phi(x,p)) } \] is minimized
Must select both metric \(\color{red}{M}\) and transformation \(\color{red}{\mathcal{T}}\)
… in addition to optimizer and the problem’s resolution
Discussed in more detail in this frontiers paper
The A-team of similarity metrics
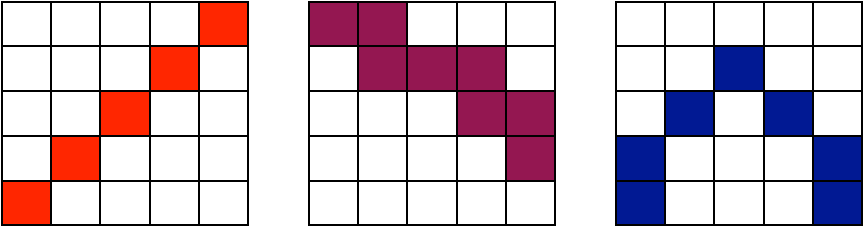
\[ \| I - J \| ~~~~~~~~~~~~~~~~~~~ \frac{< I, J >}{\|I\|\|J\|} ~~~~~~~~~~~~~~~ p(I,J) log \frac{p(I,J)}{p(I)p(J)}\]
all metrics may be computed from sparse or dense samples and used with low or high-dimensional transformations
SyN for optimization symmetry
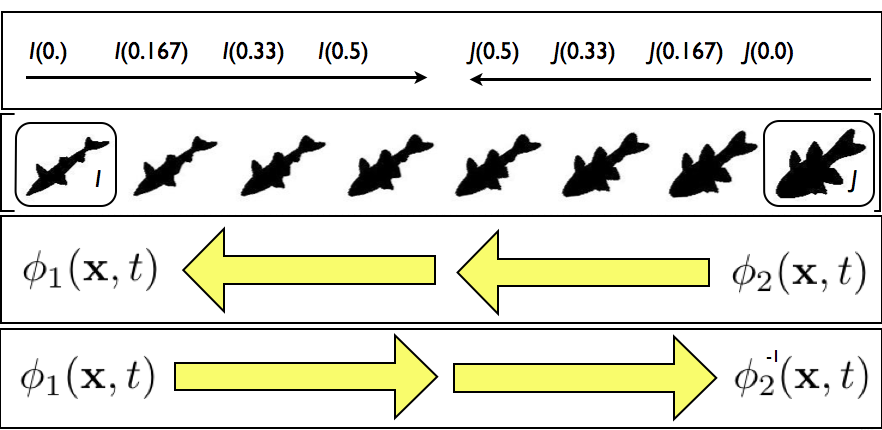 Images deform symmetrically along the shape manifold. This eliminates bias in the measurement of image differences.
Images deform symmetrically along the shape manifold. This eliminates bias in the measurement of image differences.
SyN (link) Example
Concatenated transformation \(+\) metric stages are necessary in real data
Initialize the mapping ( more on this later )
Start with a rigid transformation: \(I(x) \approx J(R(x))\) s.t. negative \(MI\) is minimized
Follow by an affine transformation: \(I(x) \approx J(R(A(x)))\) s.t. negative \(MI\) is minimized with fixed \(R\)
Finally, a diffeomorphism: \(I(x) \approx J(R(A(\phi(x))))\) s.t. \(k\)-neighborhood correlation \(CC_k\) is minimized with fixed \(R, A\)
Output the composite transform \(A \circ R\) as a matrix transformation and \(\phi\) and \(\phi^{-1}\) as deformation fields.
standard in recommended antsRegistration application scripts
Minimizing interpolations
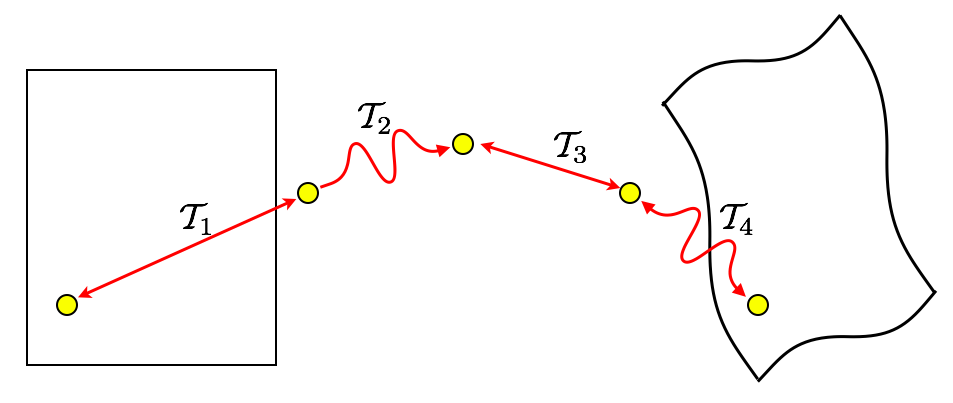
\(\mathcal{T}_{total} = \mathcal{T}_1 \circ \mathcal{T}_2 \circ \mathcal{T}_3 \circ \mathcal{T}_4\)
To avoid compounding interpolation error with the concatenation of transformations, ANTs never uses more than a single interpolation.
We ported many of these ideas into the Insight ToolKit
as part of its V4 reboot!
Registration benefits from
optimal sampling strategy
sampling for both the metric and the transformation
impacts scalability, memory, optimization accuracy, speed, robustness …
- could be done optimally with massive improvements in performance
- but needs investment in order to achieve “dream” registration scenario
important for new schemes that elect solutions from anatomical or transformation dictionaries
overall, relatively little translational work on this important problem in biomedical imaging
Sampling & feature selection: Multi-start
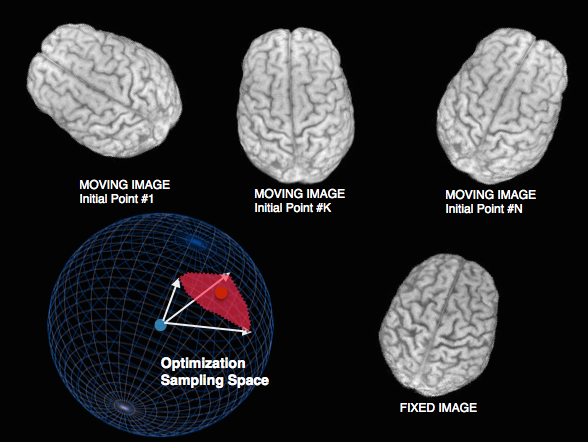
Theoretical guarantee of global optimum: improves local optimizers.
Default in antsCorticalThickness pipeline and FSL.
Sampling & feature selection: Biomedical imagery
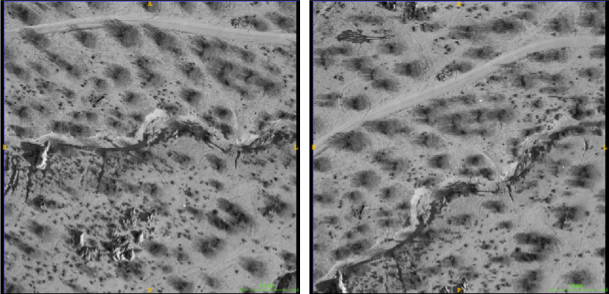
Initial configuration of data
Sampling & feature selection: Biomedical imagery
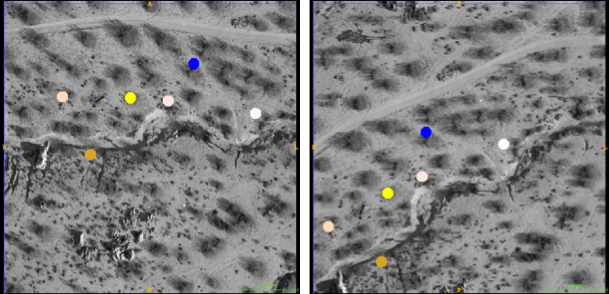
Automatic feature selection
Sampling & feature selection: Biomedical imagery
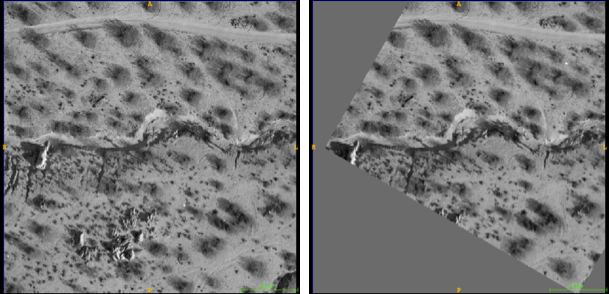
Resampling allows comparison & slide alignment and
validates the feature selection
Dramatic reduction in computation time / memory requirements
Sampling & feature selection: Lesioned brains
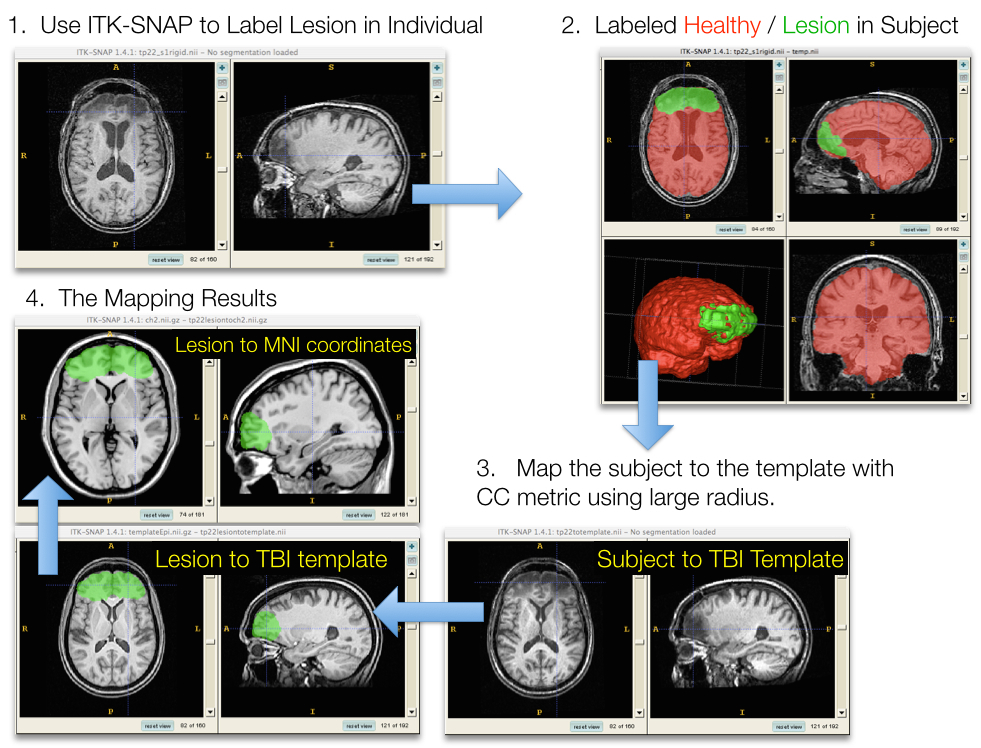
Sampling & feature selection: Summary
- we exploit these strategies to:
- accelerate
- focus
- validate
Differentiable maps with
differentiable inverse
\(+\) statistics in these spaces
Brain templates as high-dimensional averages
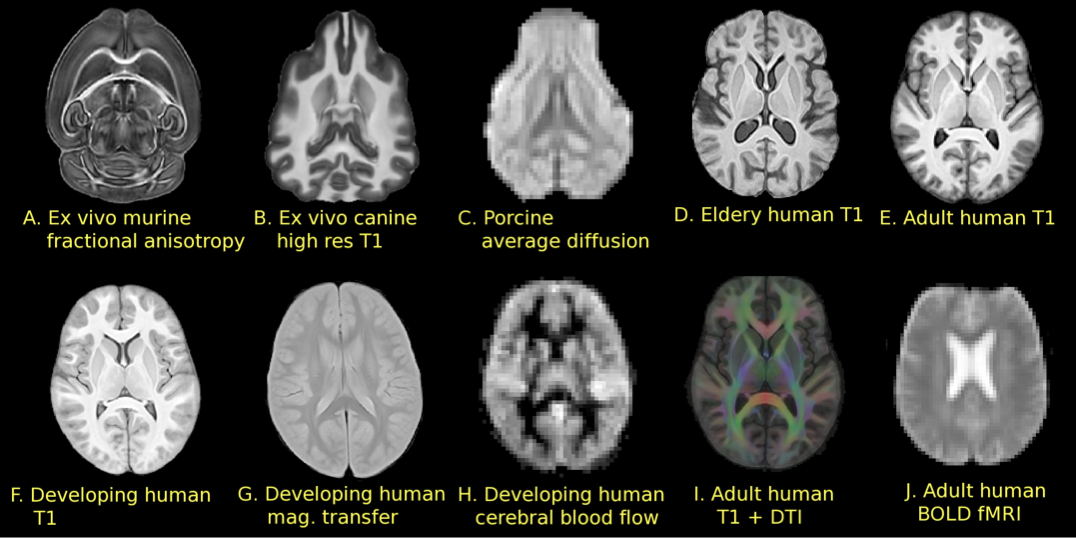
SyGN - templates and averages in deformation space
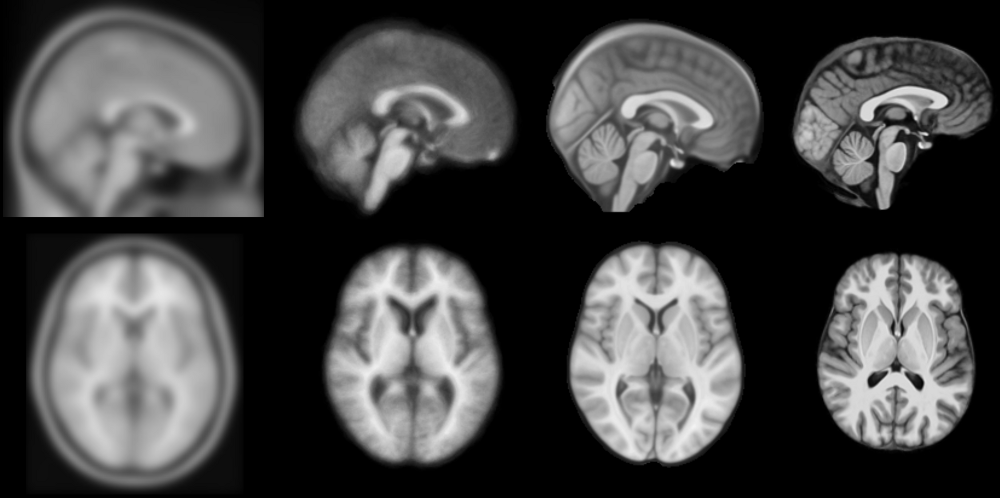 from miykael
from miykael
Statistics in deformation space
Average Republican and Democratic congressmen

We build templates to store and transfer prior knowledge
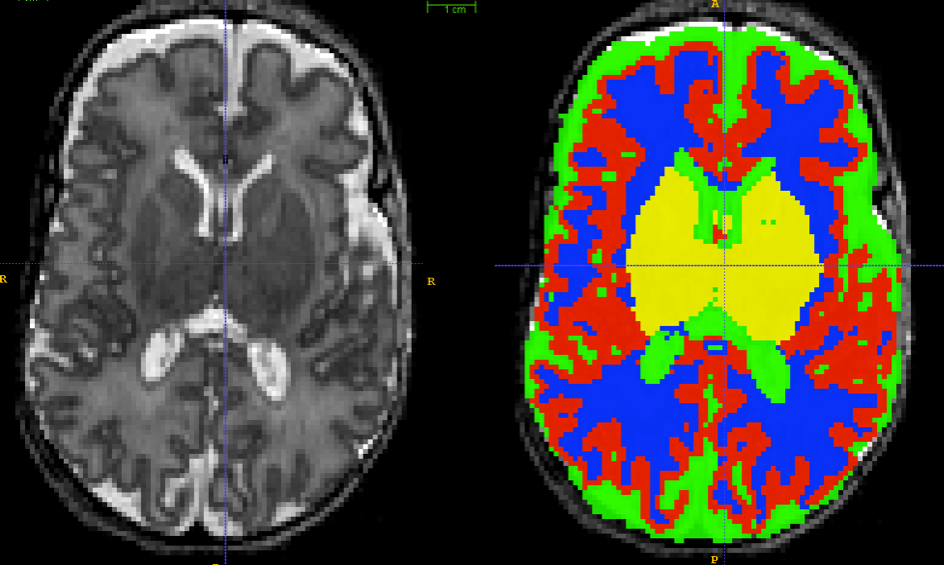
Tissue segmentation
Segmentation Framework
Bias correction (with optional priors)
Prior-based tissue segmentation
Prior-based anatomical labeling
Iteration through above steps (optional)
We tried N3 and FSL-FAST for these problems … and dislike Matlab …
failed to locate well-implemented open-source resources for general purpose prior-based segmentation and inhomogeneity correction …
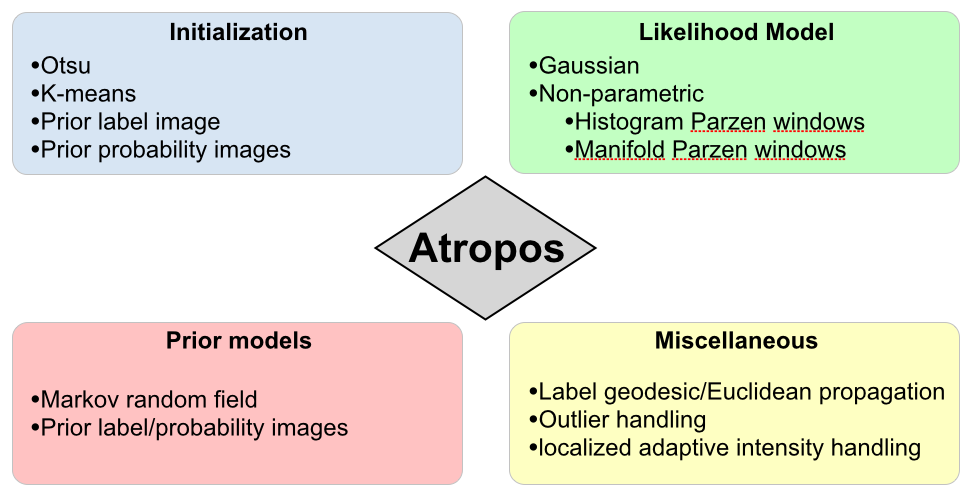
Atropos: Bayesian \(N\)-class multivariate segmentation
Similar to our experience with N3, we tried to incorporate FAST (from the FMRIB at Oxford) into an ANTs processing pipeline.
We failed to successfully incorporate priors into FAST.
Related, BA went to a segmentation-related worksop at MICCAI and aired disappointment that so much of what had been developed in the community over the last 20+ years has not been made publicly available. “What’s wrong with you people!”
3-tissue algorithm in ImageMath \(\rightarrow\) multivariate, n-class Atropos
Atropos components
Babies
Can we accurately measure cortical thickness by DiReCTly using the image space?
KellySlater \(\rightarrow\) KellyKapowski

Several years of development by SR Das, BA, NT (KK fan)
Atropos \(+\) KK Example
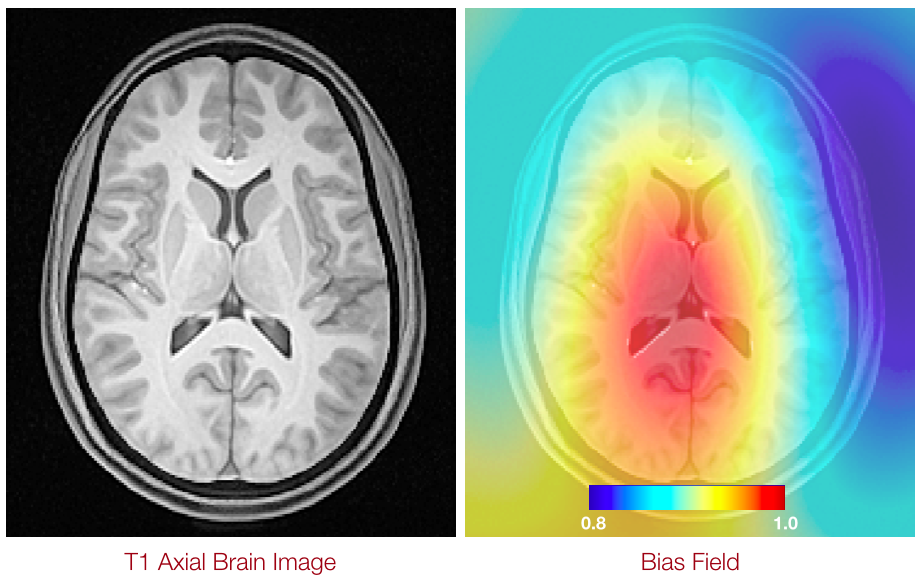
Bias Correction a.k.a. Inhomogeneity Correction
N4
N3 (developed at the Montreal Neurological Institute) has been the gold standard for bias correction—used in important projects such as ADNI
N3 is a set of perl scripts that works natively with the MINC file format which we tried to incorporate into an ANTs processing pipeline.
We had so much trouble converting back and forth between ITK-compatible Nifti format and MINC that BA suggested we try to implement N3 in ITK.
NT had some experience with B-splines and added some other tweaks giving birth to N4.
N4 Introduction
Nonparametric nonuniform intensity normalization (N3)
Sled et al., “A nonparametric method for automatic correction of intensity nonuniformity in MRI Data,” IEEE-TMI, 17(1), 1998.
Boyes et al., “Intensity non-uniformity correction using N3 on 3-T scanners with multichannel phased array coils,” NeuroImage, 39(4), 2008.
In a comparison of several correction techniques N3 performed well (Arnold et al., 2001). Also, the algorithm and software are in the public domain (http://www.bic.mni.mcgill.ca/software/N3/) and is probably the most widely used non-uniformity correction technique in neurological imaging.
Zheng et al., “Improvement of brain segmentation accuracy by optimizing non-uniformity correction using N3,” NeuroImage, 48(1), 2009.
Among existing approaches, the nonparametric non-uniformity intensity normalization method N3 (Sled et al., 1998) is one of the most frequently used… High performance and robustness have practically turned N3 into an industry standard.
Vovk et al., “A Review of Methods for Correction of Intensity Inhomogeneity in MRI,” IEEE-TMI, 26(3), 2007.
A well-known intensity inhomogeneity correction method, known as the N3 (nonparametric nonuniformity normalization), was proposed in [15]… Interestingly, no improvements have been suggested for this highly popular and successful method… The nonparametric nonuniformity normalization (N3) method [15] has obviously become the standard method against which other methods are compared.
Code
COMMAND:
N4BiasFieldCorrection
OPTIONS:
-d, --image-dimensionality 2/3/4
-i, --input-image inputImageFilename
-x, --mask-image maskImageFilename
-w, --weight-image weightImageFilename
-s, --shrink-factor 1/2/3/4/...
-c, --convergence [<numberOfIterations=50x50x50x50>,<convergenceThreshold=0.0>]
-b, --bspline-fitting [splineDistance,<splineOrder=3>]
[initialMeshResolution,<splineOrder=3>]
-t, --histogram-sharpening [<FWHM=0.15>,<wienerNoise=0.01>,<numberOfHistogramBins=200>]
-o, --output correctedImage
[correctedImage,<biasField>]
-h
--helpTalk is cheap, show me the code.
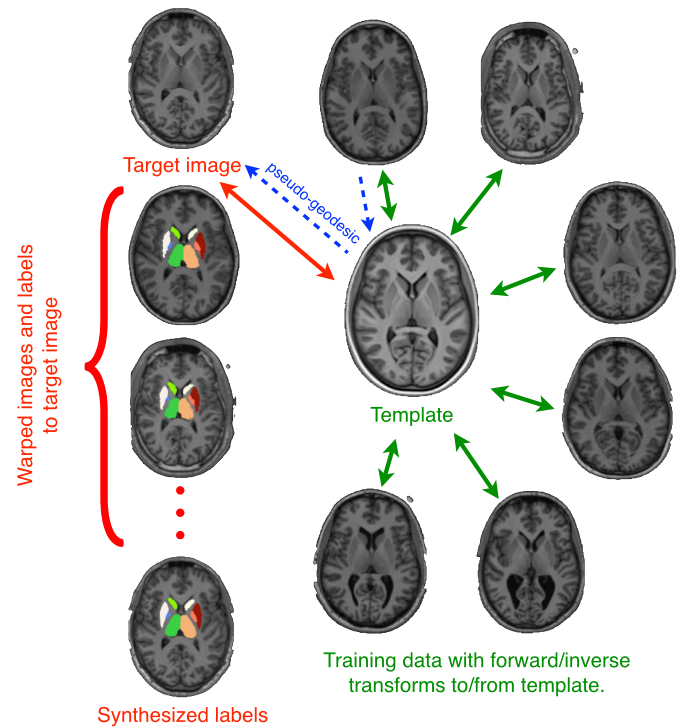
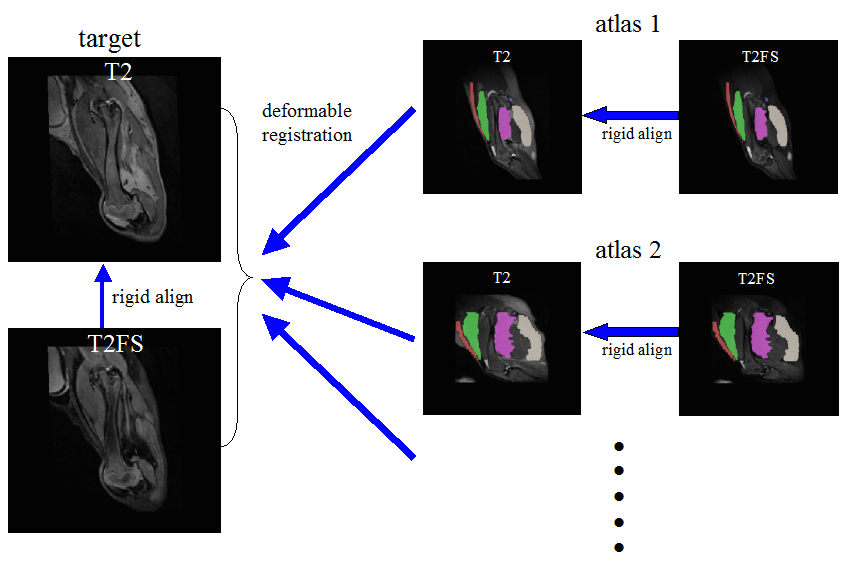
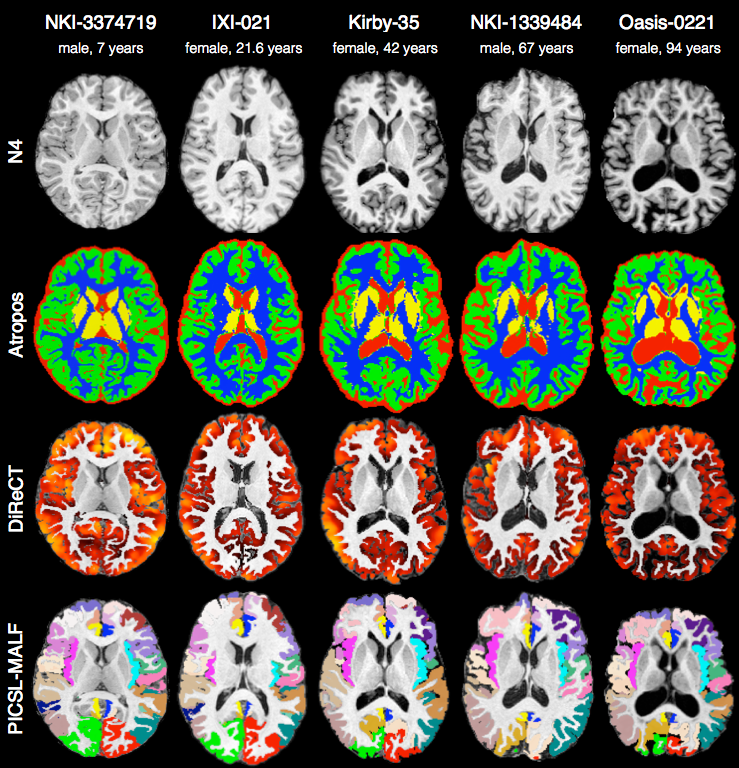
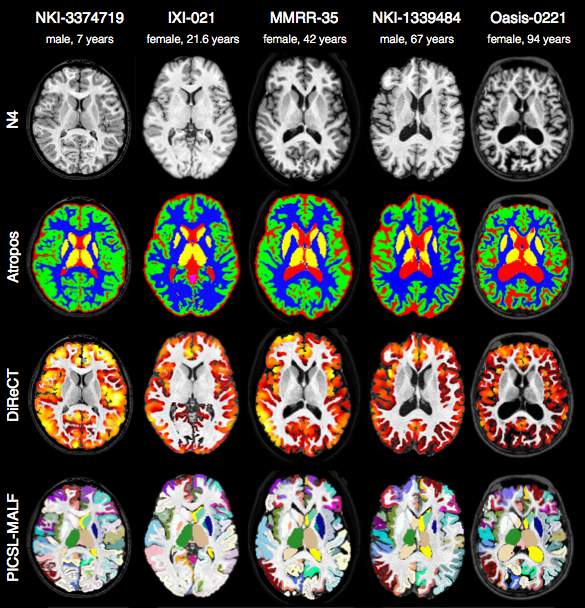
Learning from anatomical dictionaries
Joint Label Fusion
FIXME
Use dictionaries to labels
Evaluation results
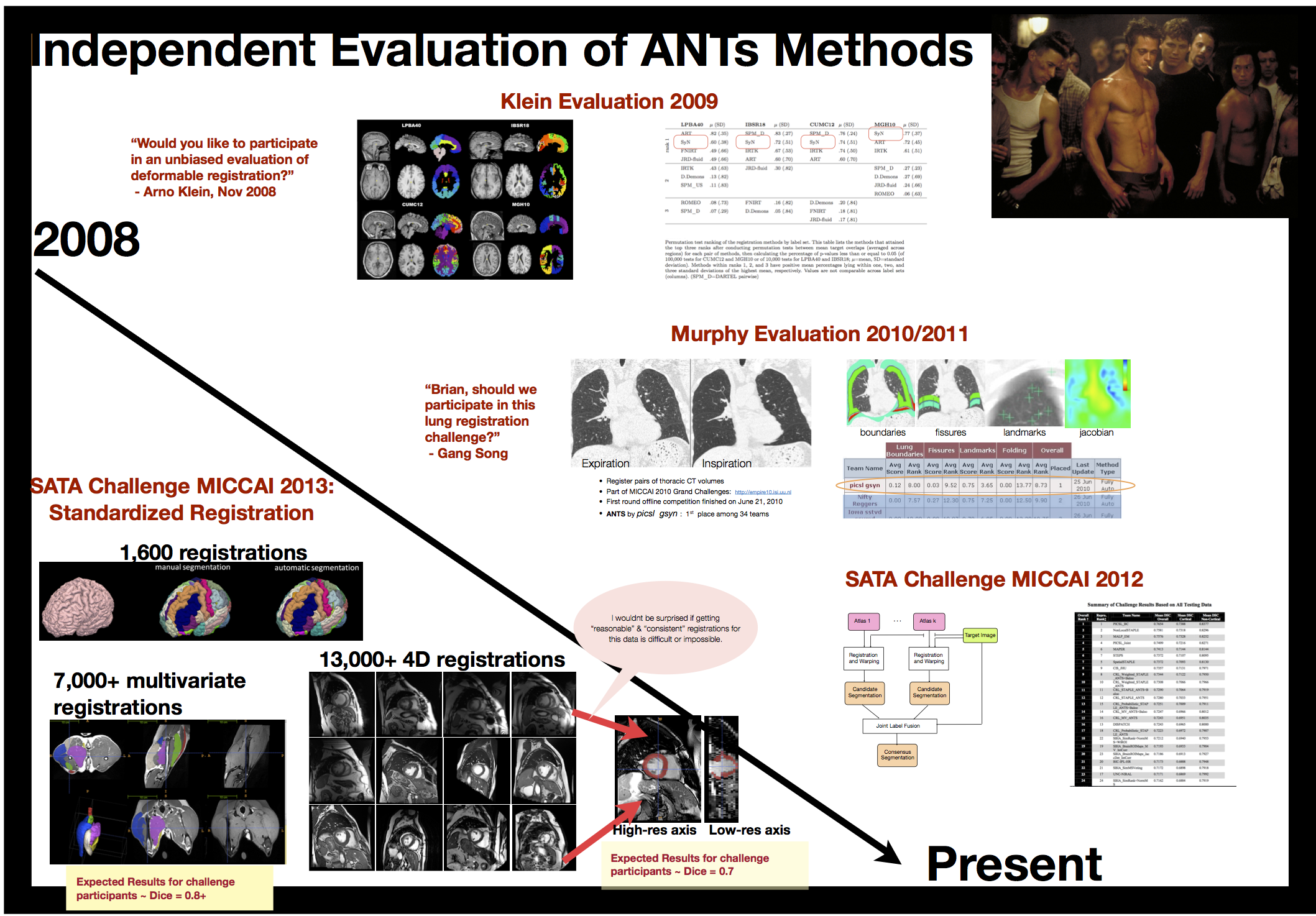
Anatomical dictionaries
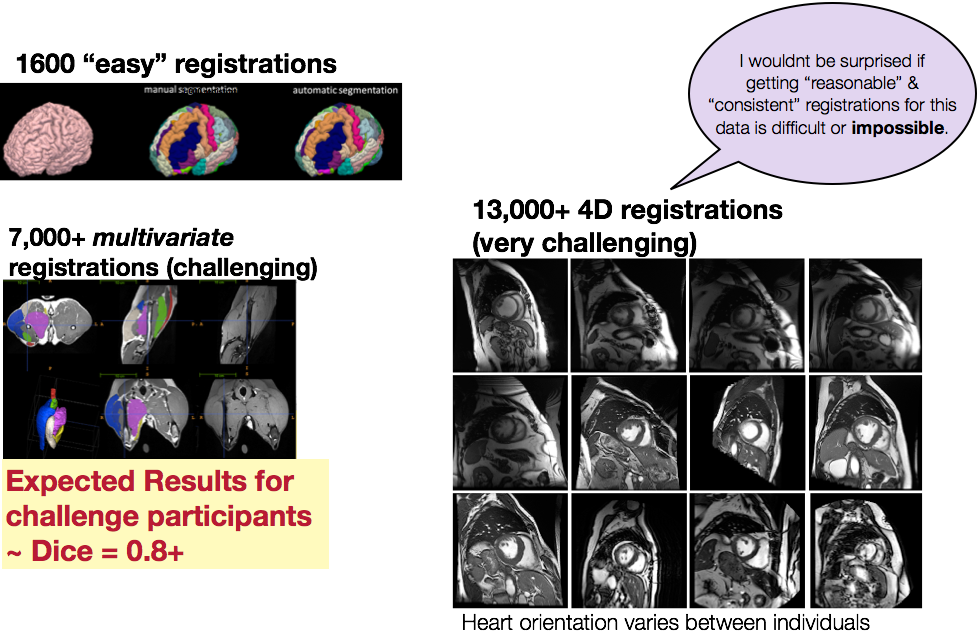
we provided the standard registration results for \(>\) 20,000 image pairs at SATA 2013
label fusion (link)
Multiple metrics improve performance
to our knowledge, ANTs is the only freely available system that can solve this problem in a fully multivariate manner.
Hongzhi Wang won the “walk in the park” award for this work …
ANTs versus Freesurfer:
Quantifying life span brain health
“Big data” problem from public resources
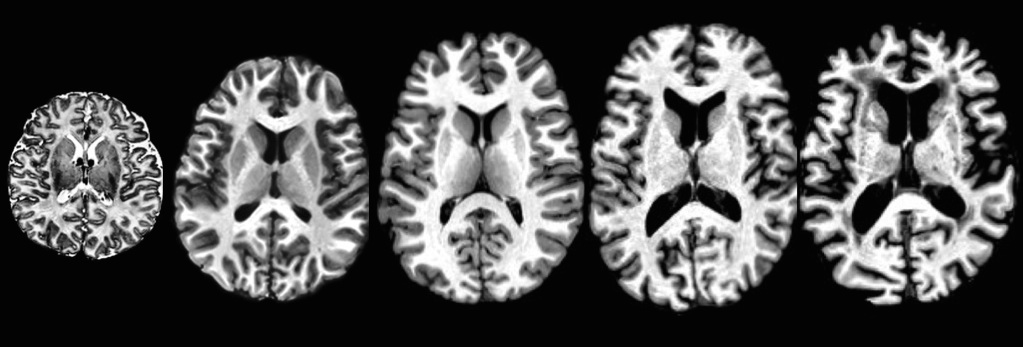
TOT, NKI, IXI, Oasis, ADNI … several thousand images
ANTs versus Freesurfer:
Quantifying life span brain health
Freesurfer is the historical standard for measuring cortical thickness
instead of using surfaces to measure cortical thickness, we use the image space DiReCTly
and this “big data” paper: Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements
comparison of prediction from automated cortical thickness measurement from 4 public datasets
\(>\) 1200 subjects, age 7 to over 90 years old
hint: ANTs thickness measurements have higher prediction accuracy relative to Freesurfer ( implying we extract more information from the data )
ANTs methods consistently improve statistical power eigenanatomy, syn, itkv4 … also, see Schwarz CG, et al. re: TBSS and related work in fMRI Miller, PNAS, Azab, et al in Hippocampus.
ANTS vs Freesurfer
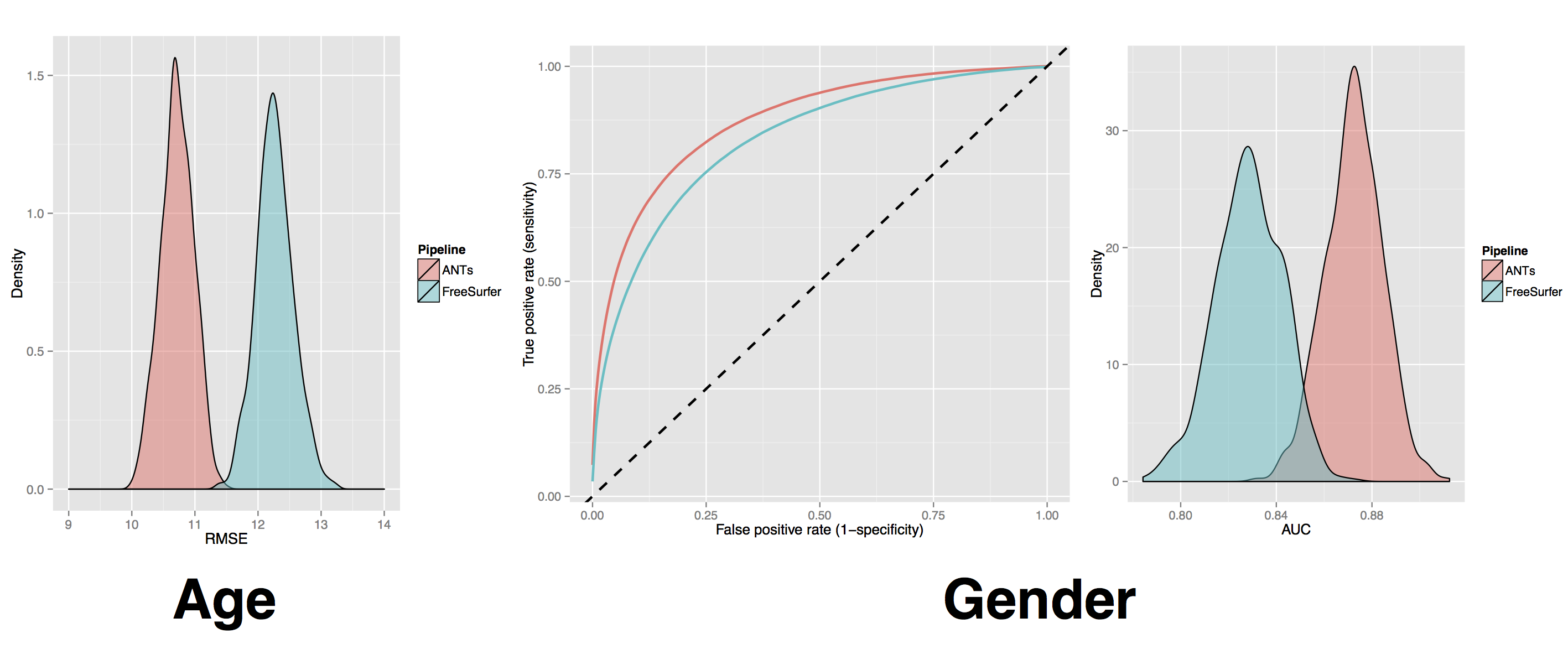
ANTs vs Freesurfer 2
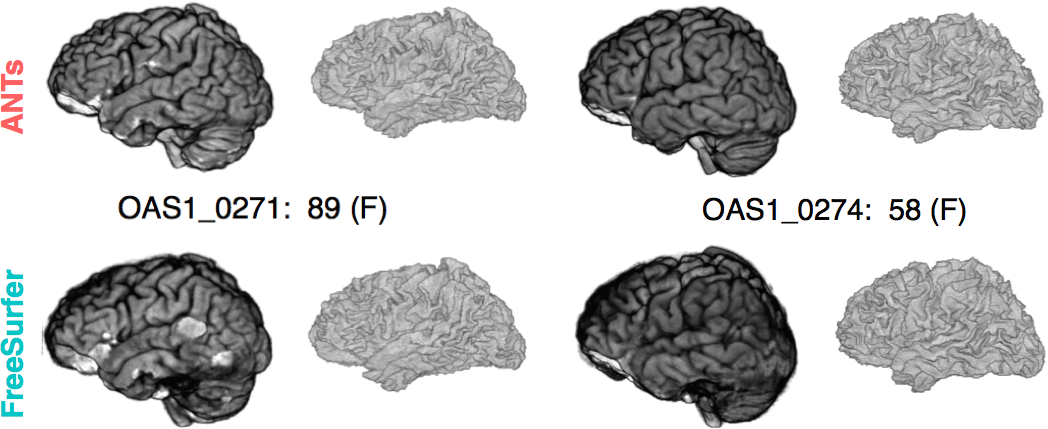
ANTs MALF Labeling
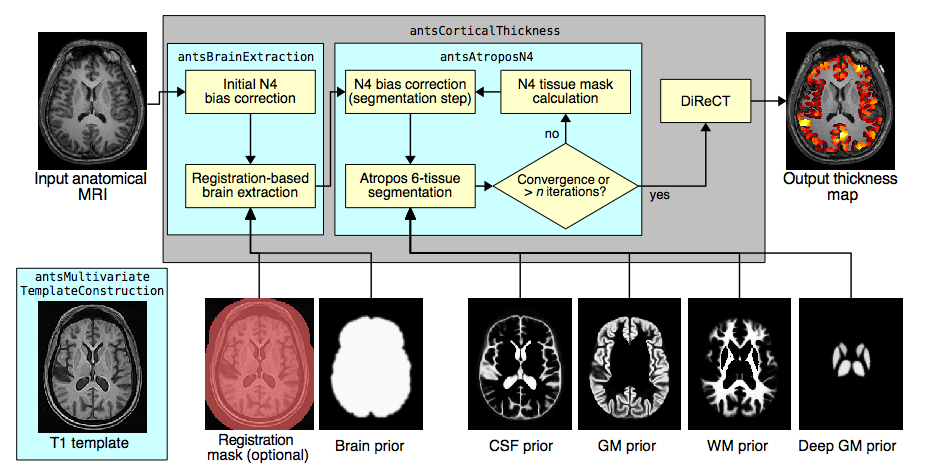
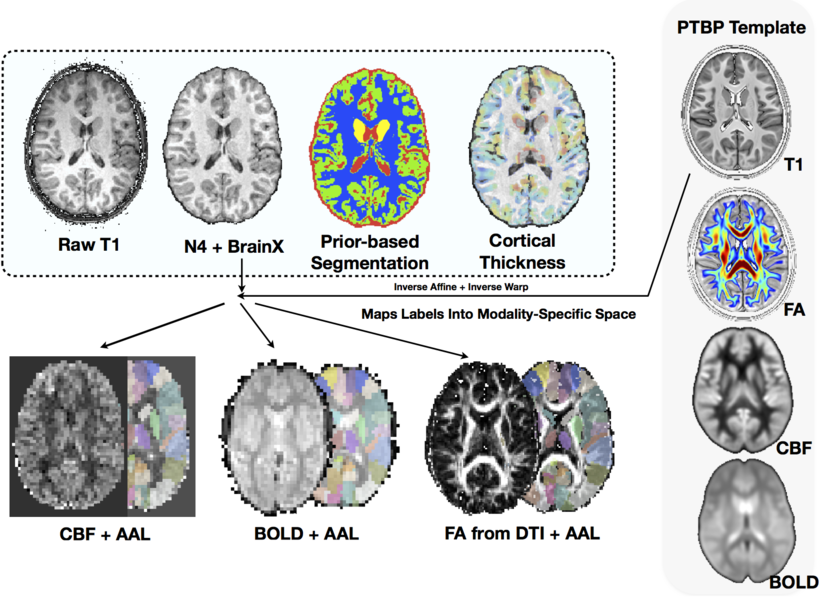
The ANTs structural brain mapping pipeline
Large-scale evaluation of ANTs* and FreeSurfer cortical thickness measurements, NeuroImage 2014.
All software components are open source and part of the Advanced Normalization Tools (ANTs) repository.
Basic components of the pipeline
- template building (offline)
- brain extraction
- cortical thickness estimation
- cortical parcellation
Template building
Tailor data to your specific cohort
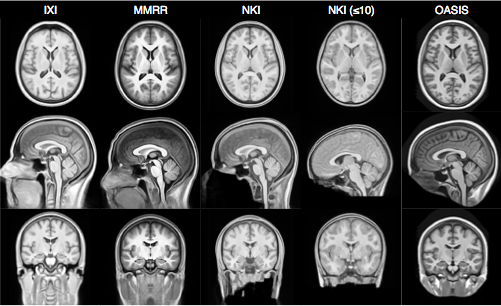
- Templates representing the average mean shape and intensity are built directly from the cohort to be analyzed, e.g. pediatric vs. middle-aged brains.
- Acquisition and anonymization (e.g. defacing) protocols are often different.
Template building (cont.)
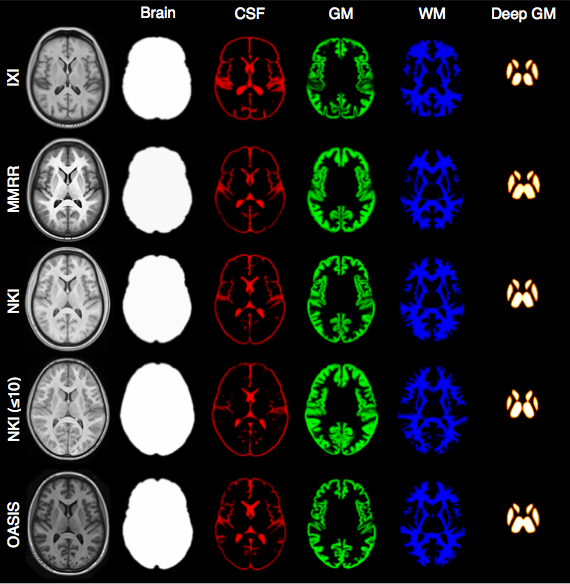
Each template is processed to produce auxiliary images which are used for brain extraction and brain segmentation.
Brain extraction
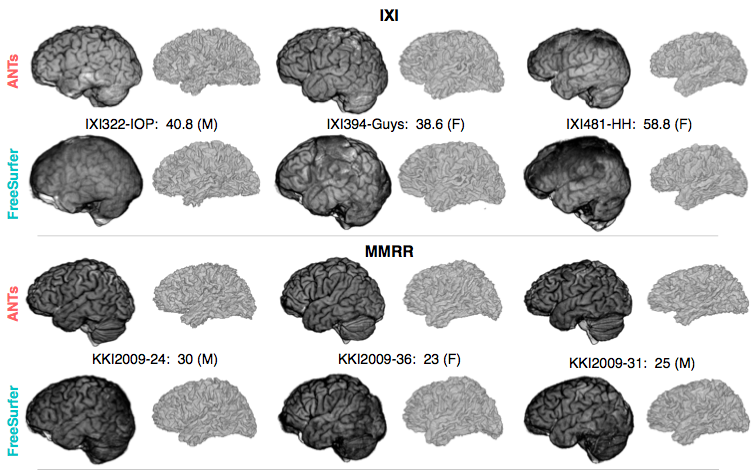
Comparison with de facto standard FreeSurfer package. Note the difference in separation of the gray matter from the surrounding CSF. (0 failures out of 1205 scans)
Brain segmentation
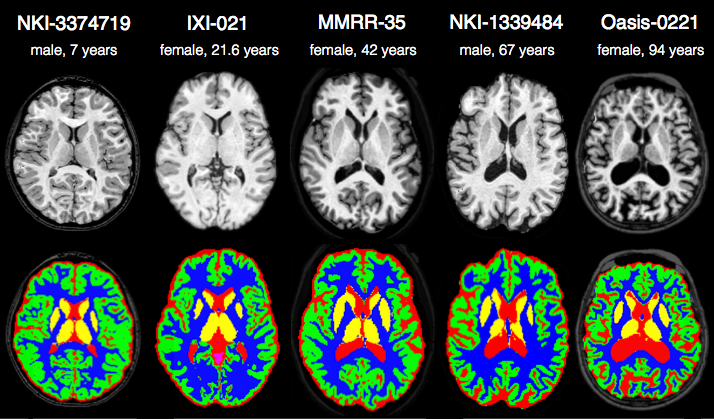
Randomly selected healthy individuals. Atropos gets good performance across ages.
Cortical thickness estimation
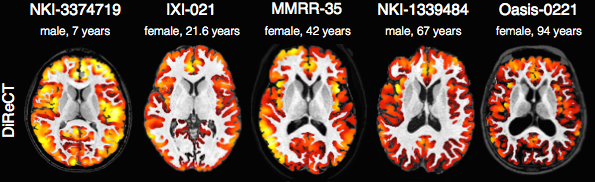
In contrast to FreeSurfer which warps coupled surface meshes to segment the gray matter, ANTs diffeomorphically registers the white matter to the combined gray/white matters while simultaneously estimating thickness.
Registration & statistics:
Frontiers and innovation
multivariate statistical fields arise from fused modalities
Many opportunities for statistical advancements
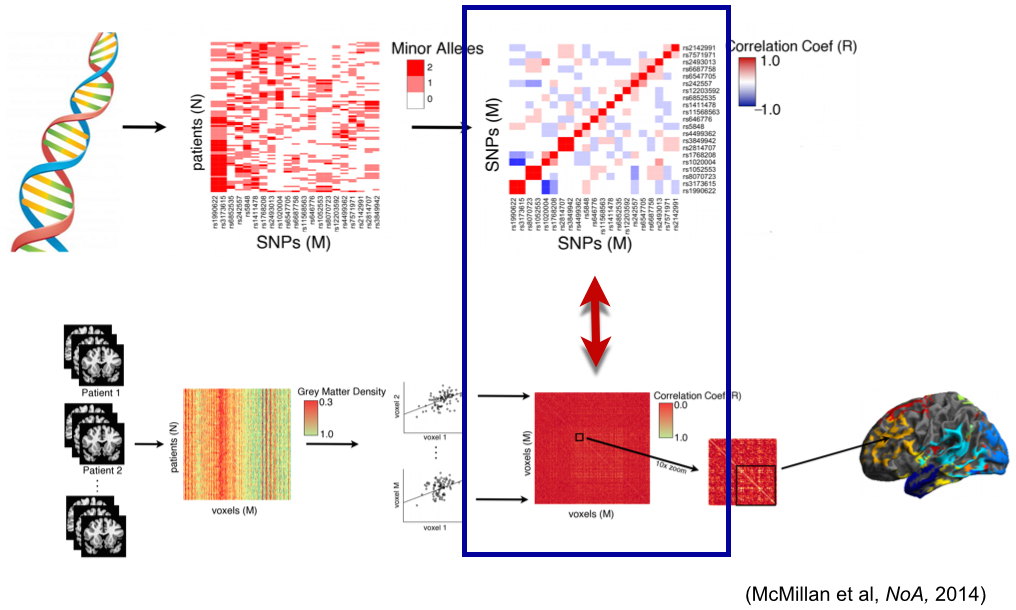
Scientific Data 2014
“Network” of predictors for age
…
ITK+ANTs+R = ANTsR
Agnostic statistics
A Quick ANTsR example
This is an executable ANTsR code block - N-dimensional statistics to go with our N-dimensional image processing software!
library(ANTsR)
dim<-2
filename<-getANTsRData('r16')
img<-antsImageRead( filename , dim )
filename<-getANTsRData('r64')
img2<-antsImageRead( filename , dim )
mask<-getMask(img,50,max(img),T)
mask2<-getMask(img,150,max(img),T)
nvox<-sum( mask == 1 )
nvox2<-sum( mask2 == 1 )The brain has 17395 voxels …
A Quick ANTsR example
Simulate a population morphometry study - a “VBM” …
simnum<-10
imglist<-list()
imglist2<-list()
for ( i in 1:simnum ) {
img1sim<-antsImageClone(img)
img1sim[ mask==1 ]<-rnorm(nvox,mean=0.5)
img1sim[ mask2==1 ]<-rnorm(nvox2,mean=2.0)
img2sim<-antsImageClone(img2)
img2sim[ mask==1 ]<-rnorm(nvox,mean=0.20)
imglist<-lappend(imglist,img1sim)
imglist2<-lappend(imglist2,img2sim)
}
imglist<-lappend( imglist, imglist2 )
mat<-imageListToMatrix( imglist, mask )
DX<-factor( c( rep(0,simnum), rep(1,simnum) ) )
mylmresults<-bigLMStats( lm( mat ~ DX ) )
qvals<-p.adjust( mylmresults$pval.model ) The minimum q-value is 2.5212 × 10-6 …
Visualize the histograms of effects
whichvox<-qvals < 1.e-2
voxdf<-data.frame( volume=c( as.numeric( mat[,whichvox] ) ), DX=DX )
ggplot(voxdf, aes(volume, fill = DX)) + geom_density(alpha = 0.2)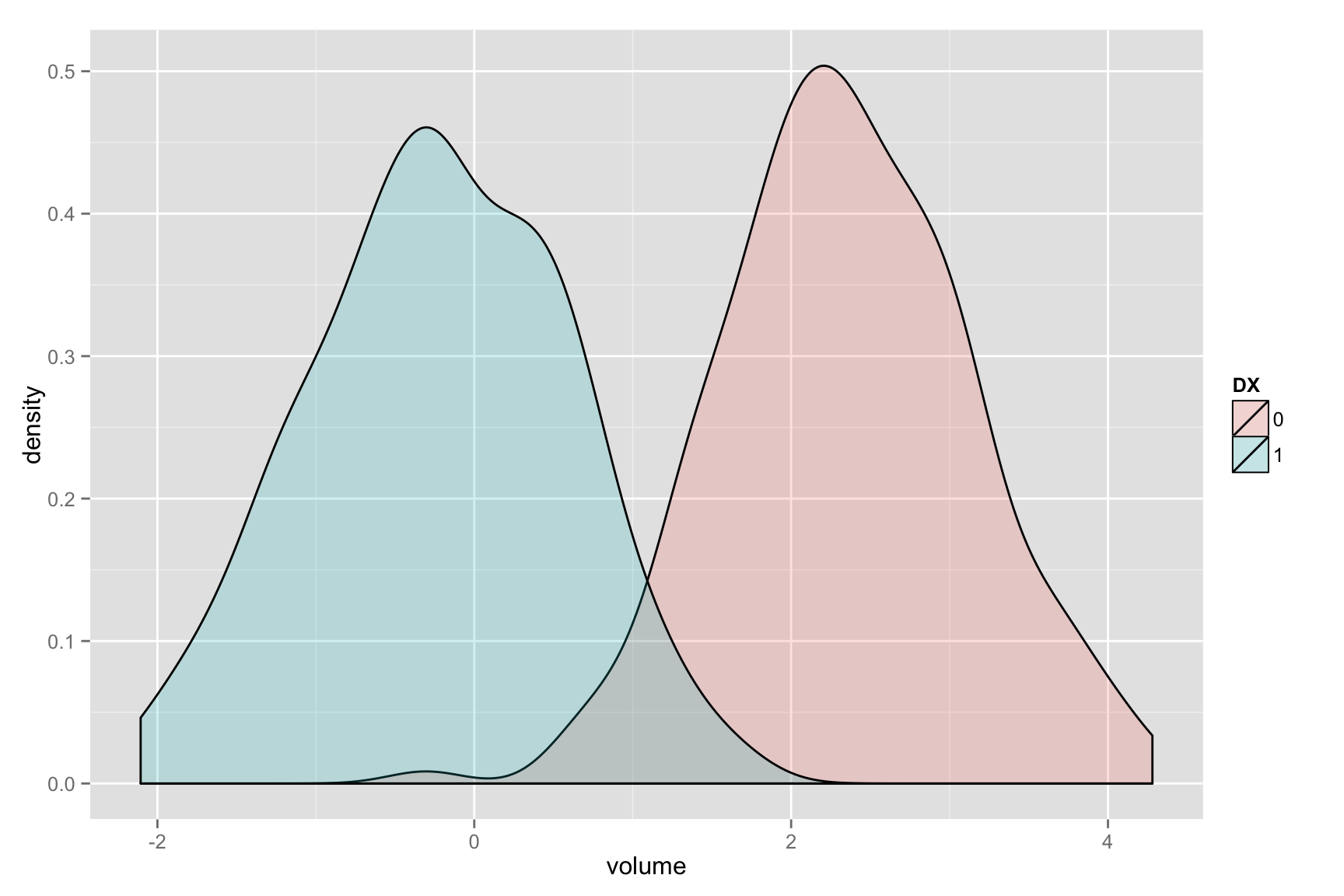
Visualize the anatomical distribution
plotANTsImage(img,functional=list(betas),threshold=thresh,
outname=ofn)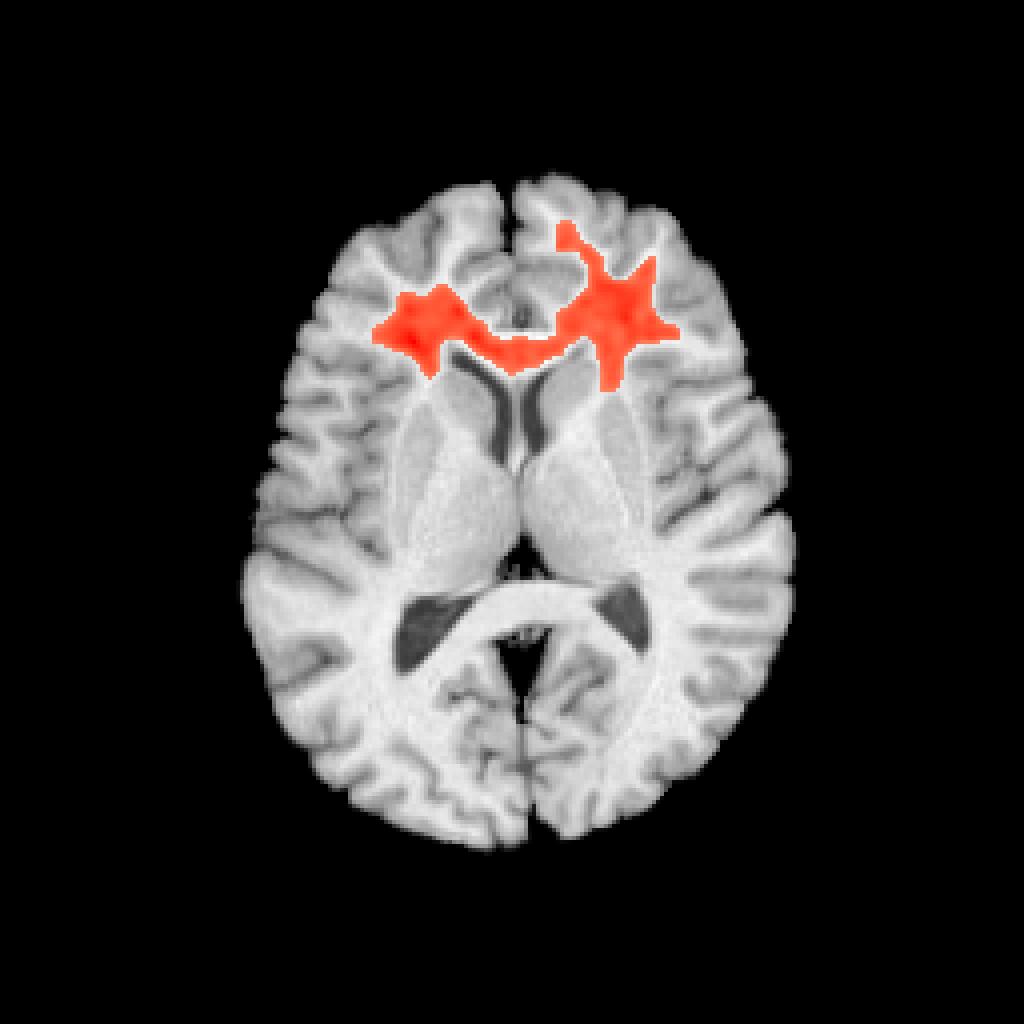
Network visualization
see ?plotBasicNetwork
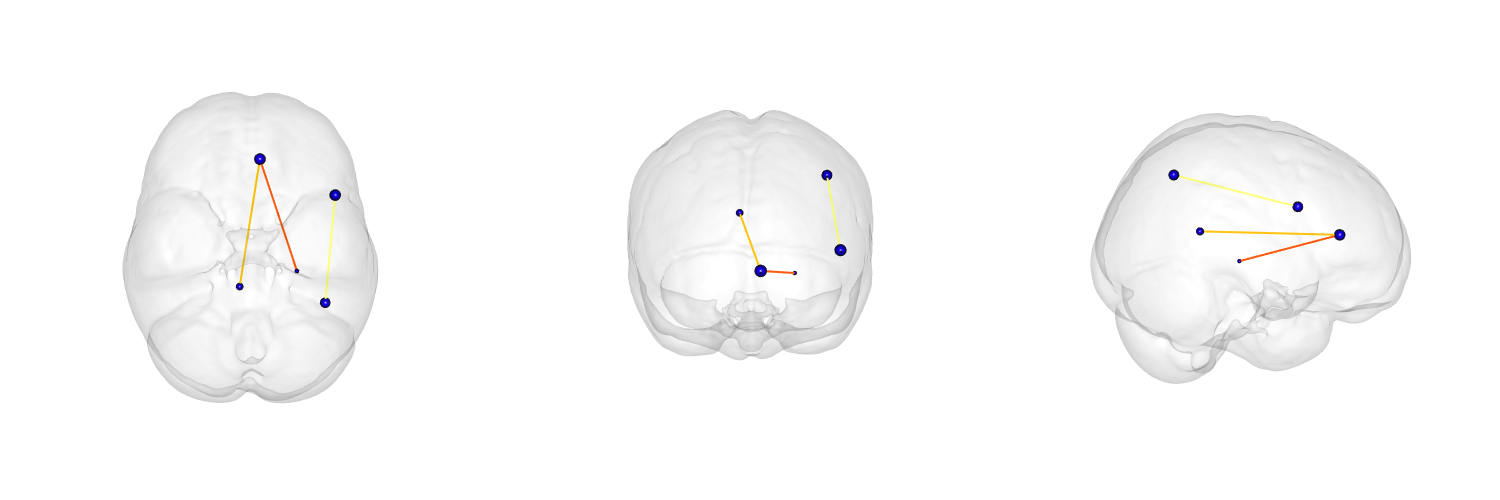
The power of ANTs \(+\) R \(\rightarrow\)
Reproducible imaging science
… used in “Sparse canonical correlation analysis relates network-level atrophy to multivariate cognitive measures in a neurodegenerative population” and several upcoming …
Wrap-up & Discussion
Many instructional examples for new colleagues
Recap
Powerful, general-purpose, well-evaluated registration and segmentation.
Differentiable maps with differentiable inverse \(+\) statistics in these spaces
Evaluated in multiple problem domains via internal studies & open competition
Borg philosophy: “best of” from I/O, to processing to statistical methods
Open source, testing, many examples, consistent style, multiple platforms, active community support …
Integration with R \(+\) novel tools for prediction, decoding, high-to-low dimensional statistics.
Collaborations with neurodebian, slicer, brainsfit, nipype, itk and more …
Challenges: Computational and Scientific
- Scalability
- need to fuse feature selection methods with transformation optimization
- need to leverage existing ITK streaming infrastructure in application level tool
- Domain expertise: Customizable for specific problems but sometimes not specific enough
- “Plausible physical modeling …” - this should vary per problem … but doesn’t.
- a fabulous project would be to resolve this issue at a large-scale e.g. for reconstructing neurons, measuring white matter elaboration …
- our prior FEM work is one potential solution
- Rapid development: colleagues still need familiarity with compilation for latest ANTs features
- Latest theoretical advances in registration not yet wrapped for users
- Need more Documentation & testing …
Tools you can use for imaging science
Core developers: B. Avants, N. Tustison, H. J. Johnson, J. T. Duda
Many contributors, including users …
Multi-platform, multi-threaded C++ stnava.github.io/ANTs
Developed in conjunction with http://www.itk.org/
R wrapping and extension stnava.github.io/ANTsR
rapid development, regular testing \(+\) many eyes \(\rightarrow\) bugs are shallow

